Biomolecules | Free Full-Text | CavitySpace: A Database of Potential Ligand Binding Sites in the Human Proteome

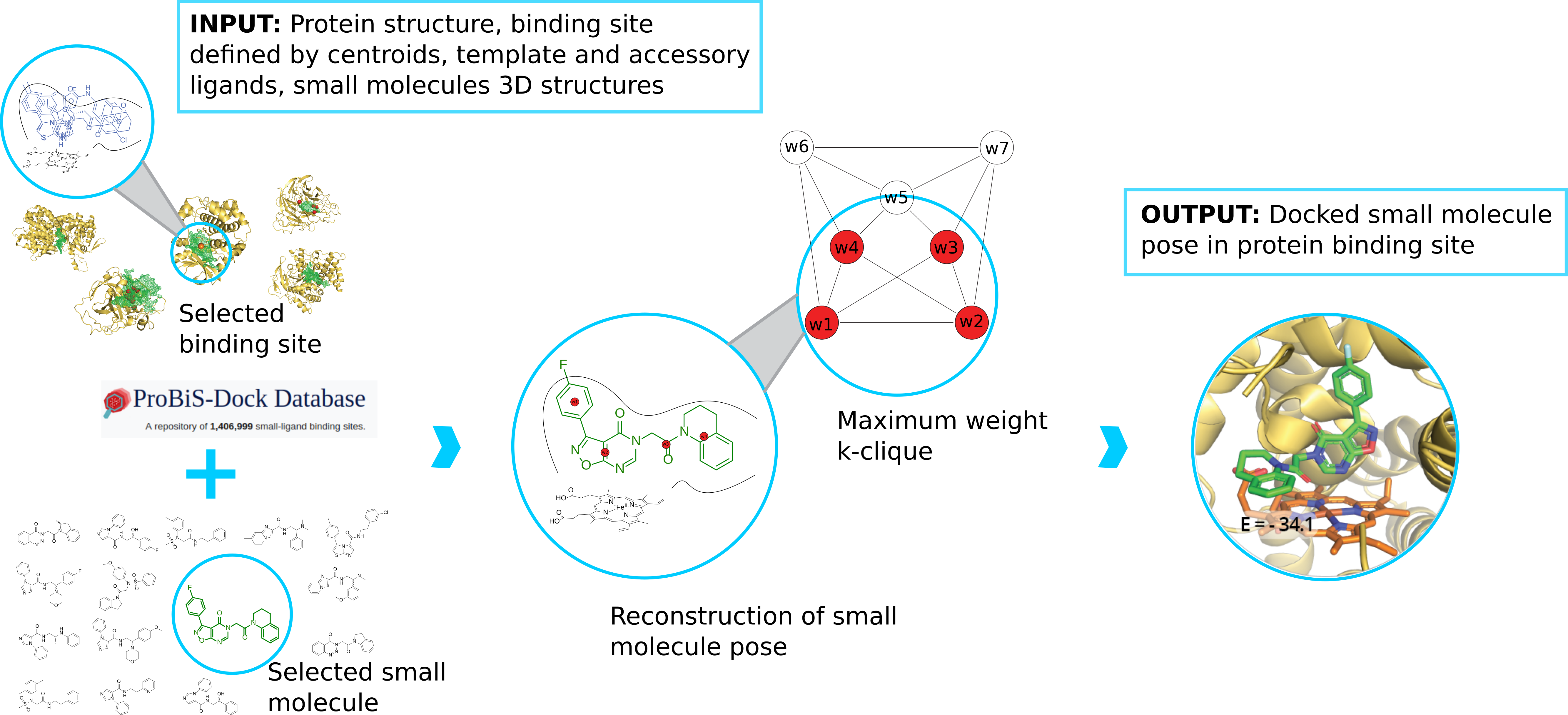
ProBiS-Dock Database: A Web Server and Interactive Web Repository of Small Ligand–Protein Binding Sites for Drug Design | Journal of Chemical Information and Modeling
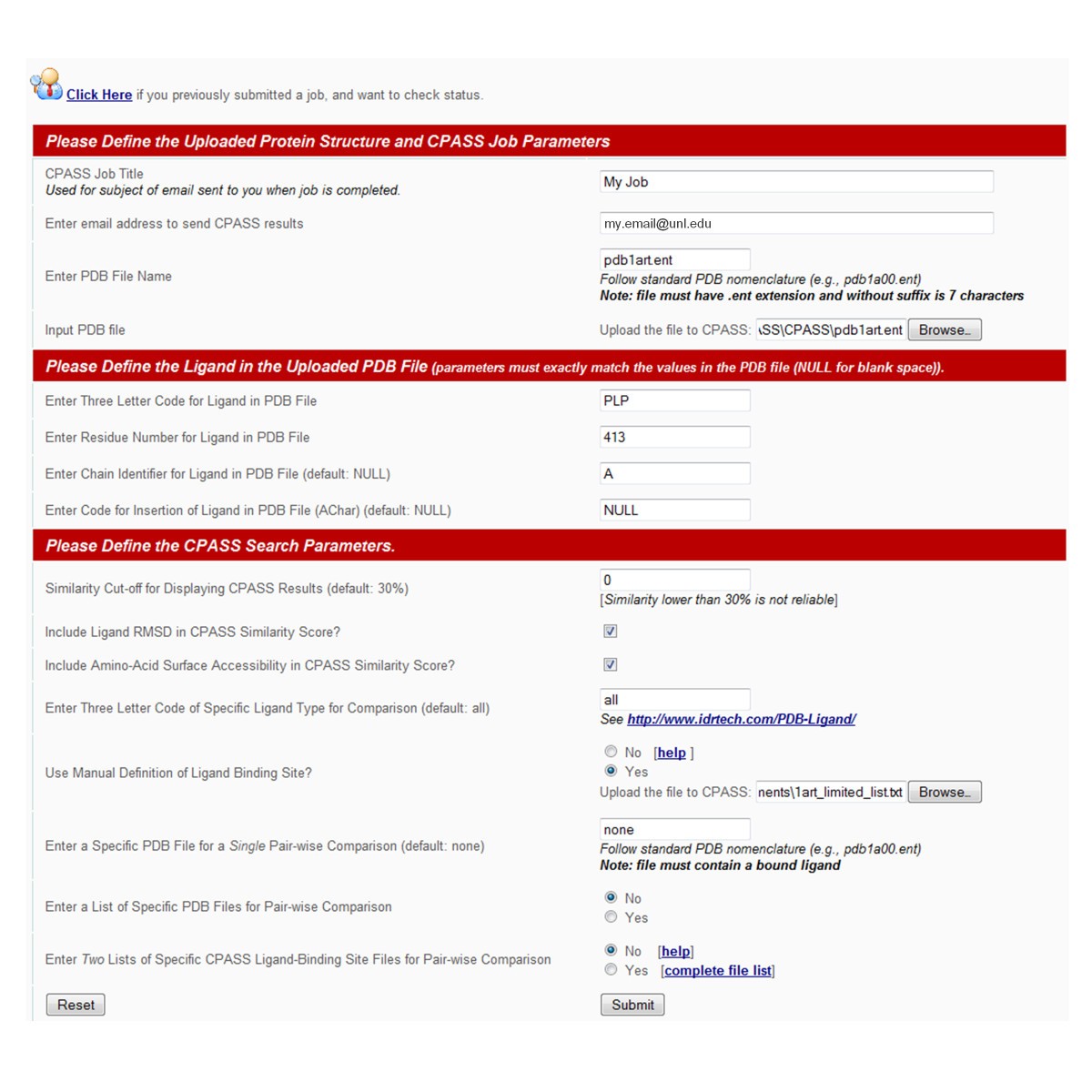
Searching the protein structure database for ligand-binding site similarities using CPASS v.2 | BMC Research Notes | Full Text
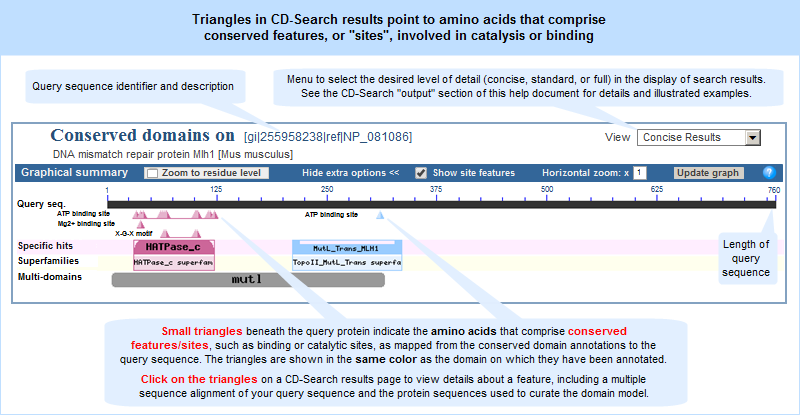
How To Use the Conserved Domain Database (CDD): identify amino acids involved in binding or catalysis

ProBiS-Fold Approach for Annotation of Human Structures from the AlphaFold Database with No Corresponding Structure in the PDB to Discover New Druggable Binding Sites | Journal of Chemical Information and Modeling

Large-Scale Analysis of Protein-Ligand Binding Sites using the Binding MOAD Database. | Semantic Scholar

ProBiS-Dock Database: A Web Server and Interactive Web Repository of Small Ligand–Protein Binding Sites for Drug Design | Journal of Chemical Information and Modeling
![PDF] sc-PDB: an Annotated Database of Druggable Binding Sites from the Protein Data Bank | Semantic Scholar PDF] sc-PDB: an Annotated Database of Druggable Binding Sites from the Protein Data Bank | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/7b4533a875fde931b28bf1906eda984350587e60/2-Figure1-1.png)
PDF] sc-PDB: an Annotated Database of Druggable Binding Sites from the Protein Data Bank | Semantic Scholar

Flowchart of the GalaxySite algorithm. The ligand-binding site of a... | Download Scientific Diagram

bSiteFinder, an improved protein-binding sites prediction server based on structural alignment: more accurate and less time-consuming | Journal of Cheminformatics | Full Text

Automatic generation of bioinformatics tools for predicting protein-ligand binding sites. - Abstract - Europe PMC

Global organization of a binding site network gives insight into evolution and structure-function relationships of proteins | Scientific Reports