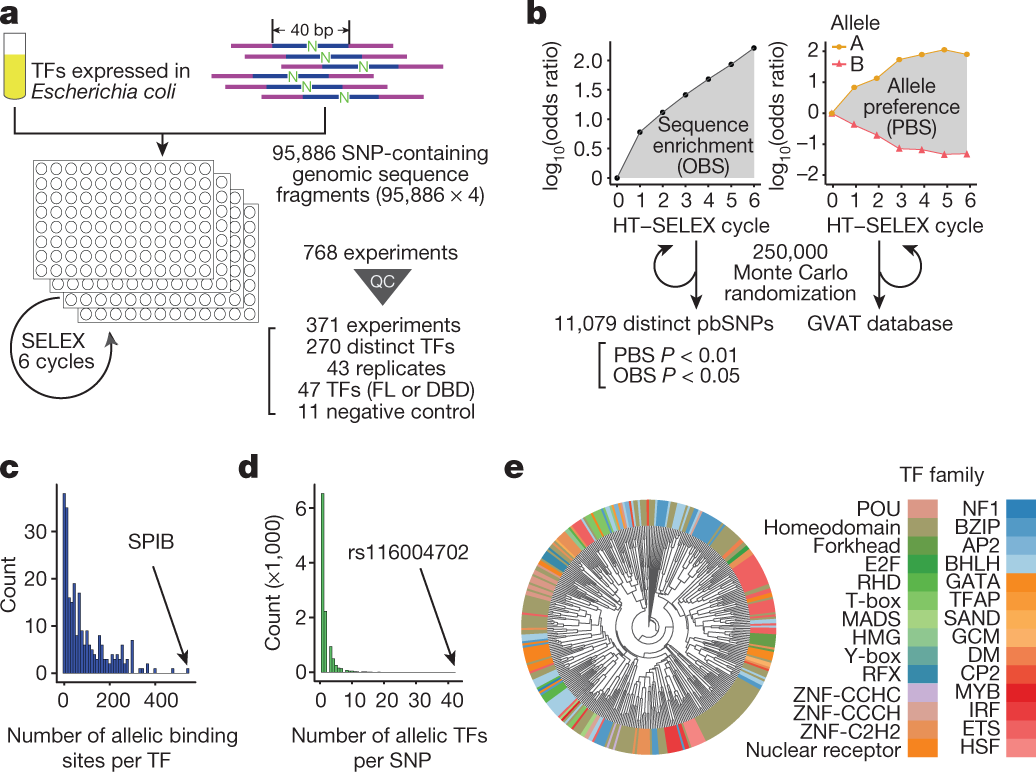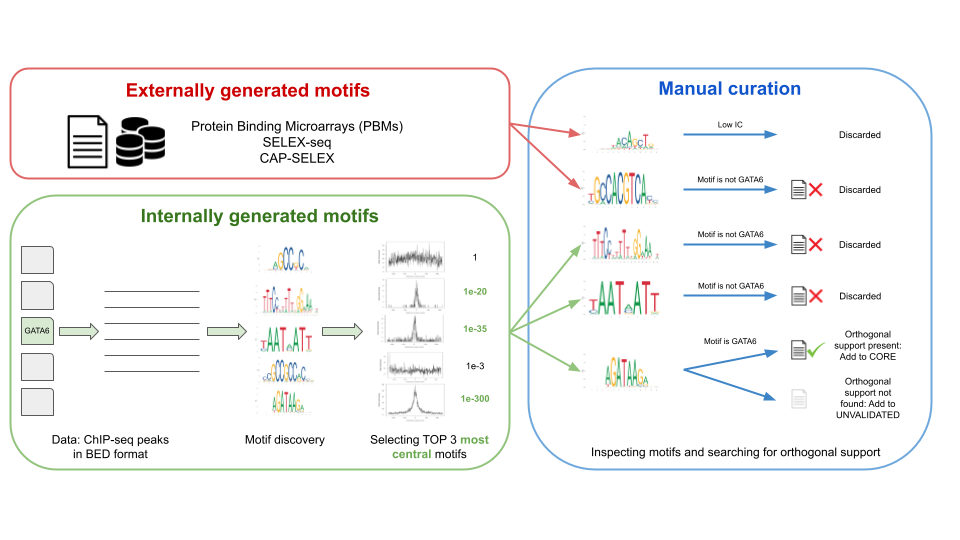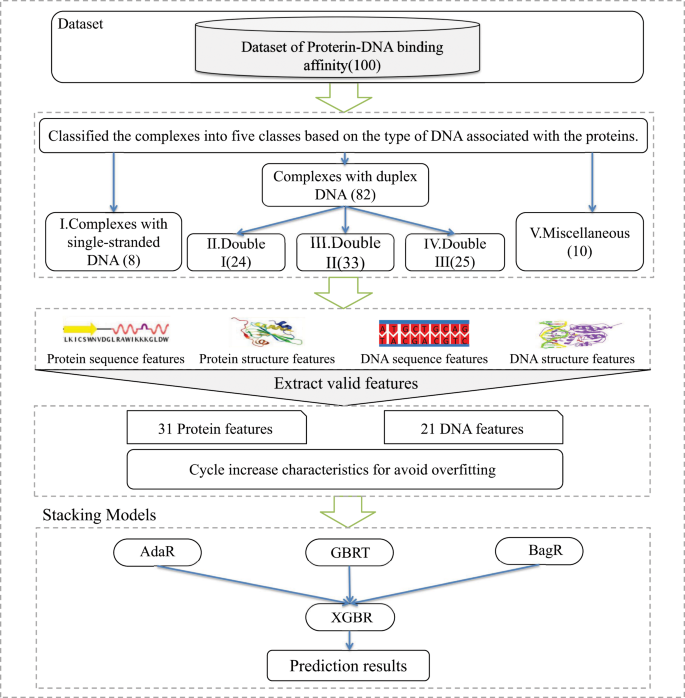
PreDBA: A heterogeneous ensemble approach for predicting protein-DNA binding affinity | Scientific Reports
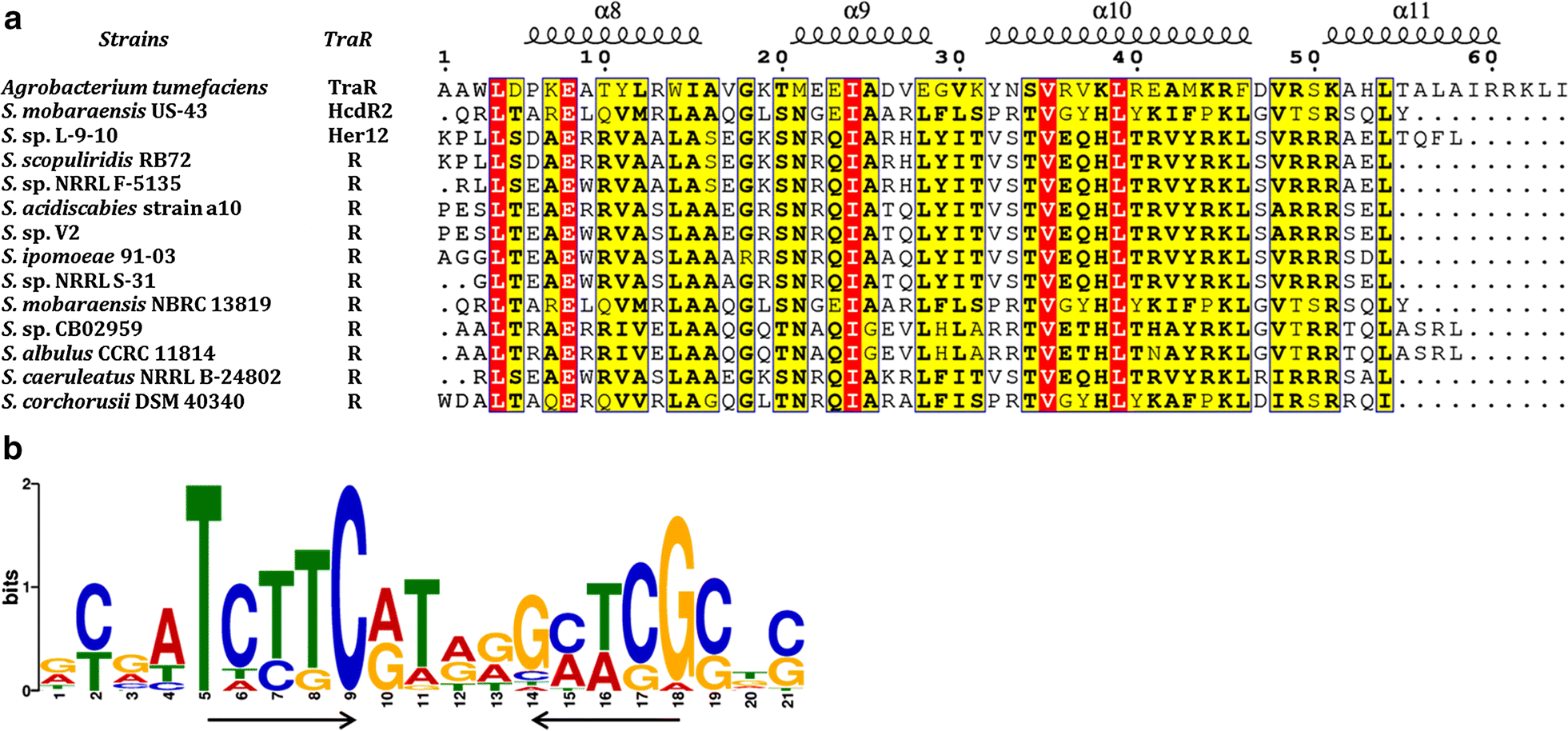
Exploring novel herbicidin analogues by transcriptional regulator overexpression and MS/MS molecular networking | Microbial Cell Factories | Full Text
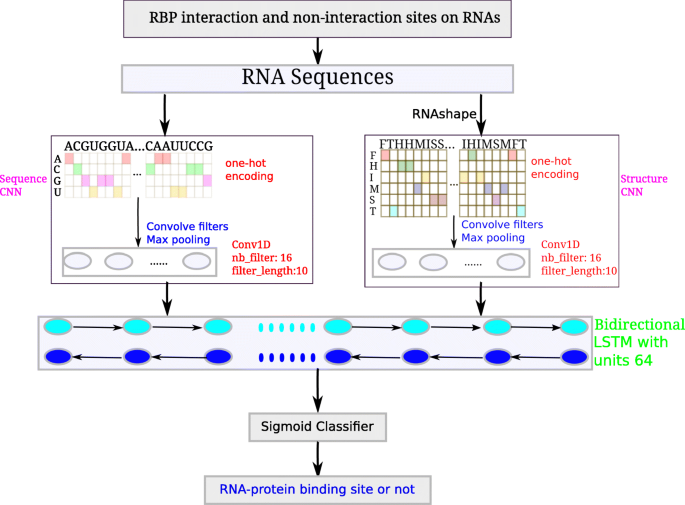
Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks | BMC Genomics | Full Text

Leopard: fast decoding cell type-specific transcription factor binding landscape at single-nucleotide resolution | bioRxiv

Improving DNA-Binding Protein Prediction Using Three-Part Sequence-Order Feature Extraction and a Deep Neural Network Algorithm | Journal of Chemical Information and Modeling

Predicting transcription factor binding sites using DNA shape features based on shared hybrid deep learning architecture: Molecular Therapy - Nucleic Acids
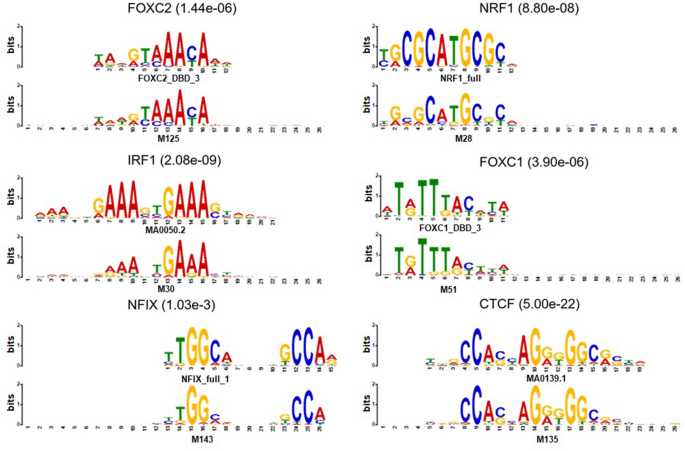
Enhancing the interpretability of transcription factor binding site prediction using attention mechanism | Scientific Reports
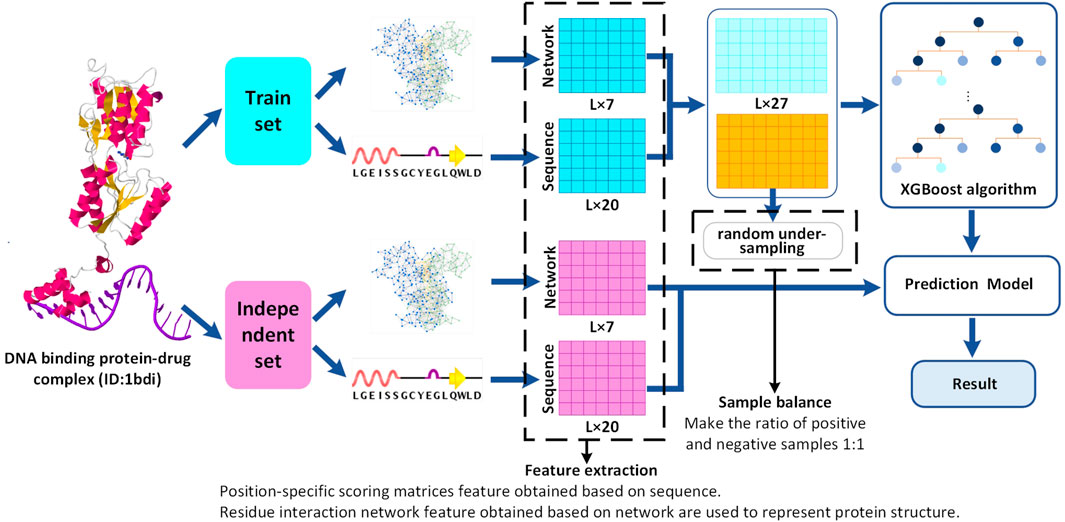
Frontiers | Prediction of DNA-Binding Protein–Drug-Binding Sites Using Residue Interaction Networks and Sequence Feature

Predicting Hot Spot Residues at Protein–DNA Binding Interfaces Based on Sequence Information | SpringerLink
![PDF] DNA Shape Features Improve Transcription Factor Binding Site Predictions In Vivo. | Semantic Scholar PDF] DNA Shape Features Improve Transcription Factor Binding Site Predictions In Vivo. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/5d68bdf0391af25147a48171a54fea49d64d16d4/2-Figure1-1.png)
PDF] DNA Shape Features Improve Transcription Factor Binding Site Predictions In Vivo. | Semantic Scholar
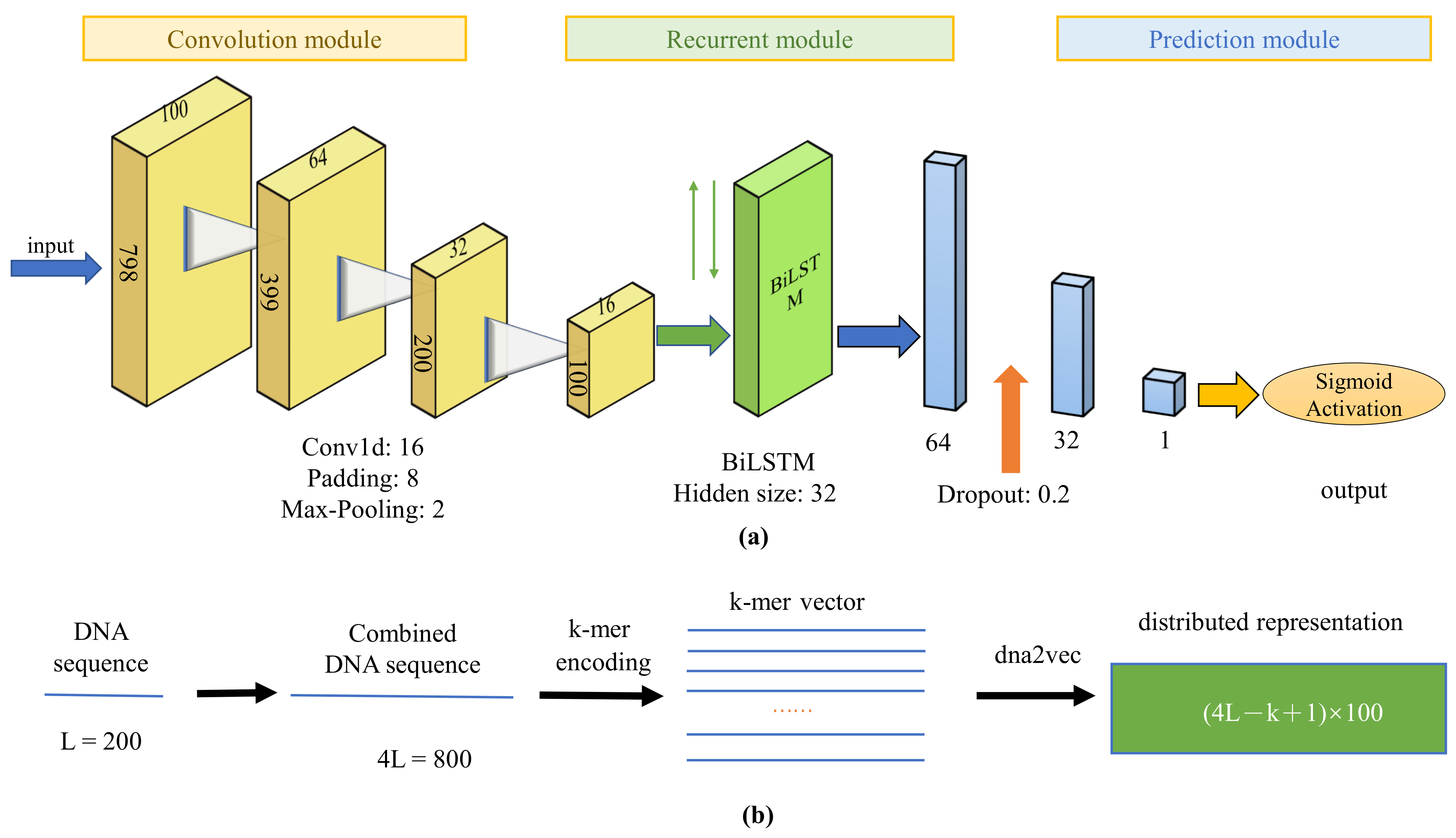
IJMS | Free Full-Text | DeepD2V: A Novel Deep Learning-Based Framework for Predicting Transcription Factor Binding Sites from Combined DNA Sequence

Prediction of transcription factor binding sites and mutation analysis... | Download Scientific Diagram
![PDF] A Review About Transcription Factor Binding Sites Prediction Based on Deep Learning | Semantic Scholar PDF] A Review About Transcription Factor Binding Sites Prediction Based on Deep Learning | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/b55033641d1547d8ca9bd969ab01d7918be338f1/6-Table2-1.png)
PDF] A Review About Transcription Factor Binding Sites Prediction Based on Deep Learning | Semantic Scholar